RES E AR C H A R T I C L E Open Access
Genome-wide identification and expression
patterns analysis of the RPD3/HDA1 gene
family in cotton
Jingjing Zhang, Aimin Wu, Hengling Wei, Pengbo Hao, Qi Zhang, Miaomiao Tian, Xu Yang, Shuaishuai Cheng,
Xiaokang Fu, Liang Ma, Hantao Wang
*
and Shuxun Yu
*
Abstract
Background: Histone deacetylases (HDACs) catalyze histone deacetylation and suppress gene transcription during
various cellular processes. Within the superfamily of HDACs, RPD3/HDA1-type HDACs are the most studied, and it is
reported that RPD3 genes play crucial roles in plant growth and physiological processes. However, there is a lack of
systematic research on the RPD3/HDA1 gene family in cotton.
Results: In this study, genome-wide analysis identified 9, 9, 18, and 18 RPD3 genes in Gossypium raimondii, G.
arboreum, G. hirsutum, and G. barbadense, respectively. This gene family was divided into 4 subfamilies through
phylogenetic analysis. The exon-intron structure and conserved motif analysis revealed high conservation in each
branch of the cotton RPD3 genes. Collinearity analysis indicated that segmental duplication was the primary driving
force during the expansion of the RPD3 gene family in cotton. There was at least one presumed cis-element related to
plant hormones in the promoter regions of all GhRPD3 genes, especially MeJA- and ABA-responsive elements, which
have more members than other hormone-relevant elements. The expression patterns showed that most GhRPD3
genes had relatively high expression levels in floral organs and performed higher expression in early-maturity cotton
compared with late-maturity cotton during flower bud differentiation. In addition, the expression of GhRPD3 genes
could be significantly induced by one or more abiotic stresses as well as exogenous application of MeJA or ABA.
Conclusions: Our findings reveal that GhRPD3 genes may be involved in flower bud differentiation and resistance to
abiotic stresses, which provides a basis for further functional verification of GhRPD3 genes in cotton development and a
foundation for breeding better early-maturity cotton cultivars in the future.
Keywords: Gossypium, Histone deacetylases, Expression patterns, Abiotic stress, Early maturity
Background
DNA combines with nuclear proteins to constitute the
chromatin, which is responsible for storing genetic and
directive information in eukaryotic cells. Chromatin is
highly arranged and mainly composed of nucleosomes,
which are formed by approximately 147 bp of DNA and
an octamer organized by the four core histone proteins_
H3, H4, H2A, and H2B [1]. Gene expression in eukary-
otes involves a complicated interaction, which is con-
trolled not only by the DNA sequence but also by
epigenetic events. Epigenetic mechanisms mainly consist
of histone modification and DNA methylation, and play
an important role in the regulation of gene expression.
In general, histone posttranslational modifications, in-
cluding methylation, acetylation, phosphorylation, ADP-
ribosylation an d ubiquitination, occur at the N-terminal
of histones [2], and these changes facilitate the binding
© The Author(s). 2020 Open Access This article is licensed under a Creative Commons Attribution 4.0 International License,
which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give
appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if
changes were made. The images or other third party material in this article are included in the article's Creative Commons
licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons
licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain
permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the
data made available in this article, unless otherwise stated in a credit line to the data.
State Key Laboratory of Cotton Biology, Institute of Cotton Research of
Chinese Academy of Agricultural Sciences, Anyang 455000, Henan, China
Zhang et al. BMC Genomics (2020) 21:643
https://doi.org/10.1186/s12864-020-07069-w
of other proteins to DNA, resulting in synergistic or an-
tagonistic regulation of gene transcription [3, 4] . Among
the several histone modifications, histone acetylation is a
reversible process that plays essential roles in epigenetic
regulation. The acetyla tion of core histones is catalyzed
by histone acetyltransferases (HATs) to promote tran-
scriptional activation, whereas deacetylation is regulated
by histone deacetylases (HDACs) that drive the tran-
scriptional suppression [5]. HDACs deacetylate the ly-
sine residues of N-terminal histone tails, resulting in the
repression of gene expression [6].
HDACs are involved in a large amount of biological
processes associated with plant growth and development
[7–9]. Based on sequence homology to yeas t HDACs,
HDACs in plants are divided into three main categories:
reduced potassium dependency 3 / histone deacetylase 1
(RPD3/HDA1), histone deacetylase 2 (HD2), and silent
information regulator 2 (SIR2)[7, 10, 11]. RPD3/HDA1-
type histone deacetylases, which are homologous to
yeast RPD3 and HDA-1, belong to a large family, and
they require zinc ions to catalyze activity; the HDAC in-
hibitor trichostatin A (TSA) or sodium butyrate can in-
hibit their enzymatic activities [7]. The Arabidopsis
RPD3/HDA1 gene family is further classified into three
groups. Class I includes HDA6, HDA7, HDA9, and
HDA19; class II includes HDA5, HDA15, and HDA18;
and HDA2 is the only member of class III [7 , 8]. The
other genes of PRD3/HDA1 family are unclassified in
Arabidopsis.
Over the past 20 years, RPD3/HDA1-type HDACs (call
RPD3 for short below) have been studied extensively as
global regulatory factors playing essential roles in a
series of plant growth and development processes and
the response to various environmental stresses [8, 12–
14]. In Arabidopsis, it has been reported that AtHDA19
was involved in various developmental processes, includ-
ing flowering time, circadian clocks functions, and seed
development [15,
16]. Additionally, AtHDA19 might
regulate gene expression related to jasmonic acid and
ethylene signaling pathways in response to wounding
and path ogen infection [17]. In maize, the expression
patterns of the three ZmPRD3 genes ZmRpd3/101,
ZmRpd3/102, and ZmRpd3/108 showed widespread ex-
pression in all investigated corn organs. Furthermore,
the gene products could be detected in all cellular parts
at specific stages such as kernel, shoot, and anther devel-
opmental periods [18]. In rice, HDA705 responded to
ABA and abiotic stresses, and its expression was induced
by JA. In addition, the expression of HDA702 and
HDA704 was significantly induced by SA, JA, or ABA
[19, 20]. These findings indicate that the RPD3 members
play an important regulatory role in plant development
and in the response to various stresses and plant
hormones.
Cotton is one of the most important economic crops
in China with an essential role in the national economy.
Early maturity and stress resistance are vital target traits
of cotton breeding. Over the past two decades, the RPD3
genes have been intensively studied, and some progress
has been made in Arabidopsis and some other crops.
However, there is a lack of systematic research on the
RPD3 gene family in cotton. Thus, it is necessary to ex-
plore the potential functions of RPD3 genes in cotton. In
our study, the protein seq uences of cotton RPD3-type
HDACs were predicted by genome-wide identification
and the phylogenetic tree, gene structure, conserved
motif, protein do main, expression profiles, and prelimin -
ary functions were comprehensively analyzed. The infor-
mation gained for GhRPD3 provides a reference for
further exploration of the possible functions of RPD3
genes in cotton growth and development.
Results
Identification of RPD3 genes in nine species
In this study, a total of 108 RPD3 protein sequences
from nine species were identified after eliminating re-
dundant sequences, and they are named by the pos-
ition on the chromosome. The corresponding
relationship be tween gene ID n umber and gene name
is shown in Additional file 1:TableS1.Atotalof18
genes (GhHDA1-GhHDA18) containing Hist_deacetyl
(PF00850) domains w ere identified from G. hirsutum;
9 genes were located on the A t genome, and 9 genes
were mapped on the Dt genome. Furthermore, 18
genes (GbHDA1-GbHDA18)fromG. barbadense,9
genes (GaHDA1-GaHDA9)fromG. aboreum,and9
genes ( GrHDA1-GrHDA 9)fromG. raimondii were
detected. Tetraploid c otton possessed twice as many
RPD3 genes as diploid cotton, indicating that no
RPD3 cotton gene was lost in t he process of poly-
ploidy. The numbers of RPD3 genes in the other five
species were 10 (Arabidopsi s), 14 (Oryza sativa L.),
11 (Populus trichocarpa), 8 (Theobroma cacao), and
11 (Zea mays L.). The GhRPD3 protein length ranged
from 232 to 635 aa with an a verage o f 459 aa. The
physicochemical parameters showed that the isoelec-
tricpoint(pl)ofGhRPD3 proteins varied from 4.47
to 8.65 with an average value of 5.68, and the mo-
lecular w eight of GhRPD3 proteins varied f rom 25.79
to 73.01 kDa with an avera ge value of 51.21 k Da. The
subcellular localization results indicated that most of
the GhRPD3 genes were located in cytoplasmic (10)
or nuclear (8), suggesting that GhRPD3 genes might
possess multiple regulatory functions (Table 1). The
predicted length, pI, MW and subcellular localization
of the RPD3 p roteins in other eight species are shown
in Additional file 1: Table S1.
Zhang et al. BMC Genomics (2020) 21:643 Page 2 of 16

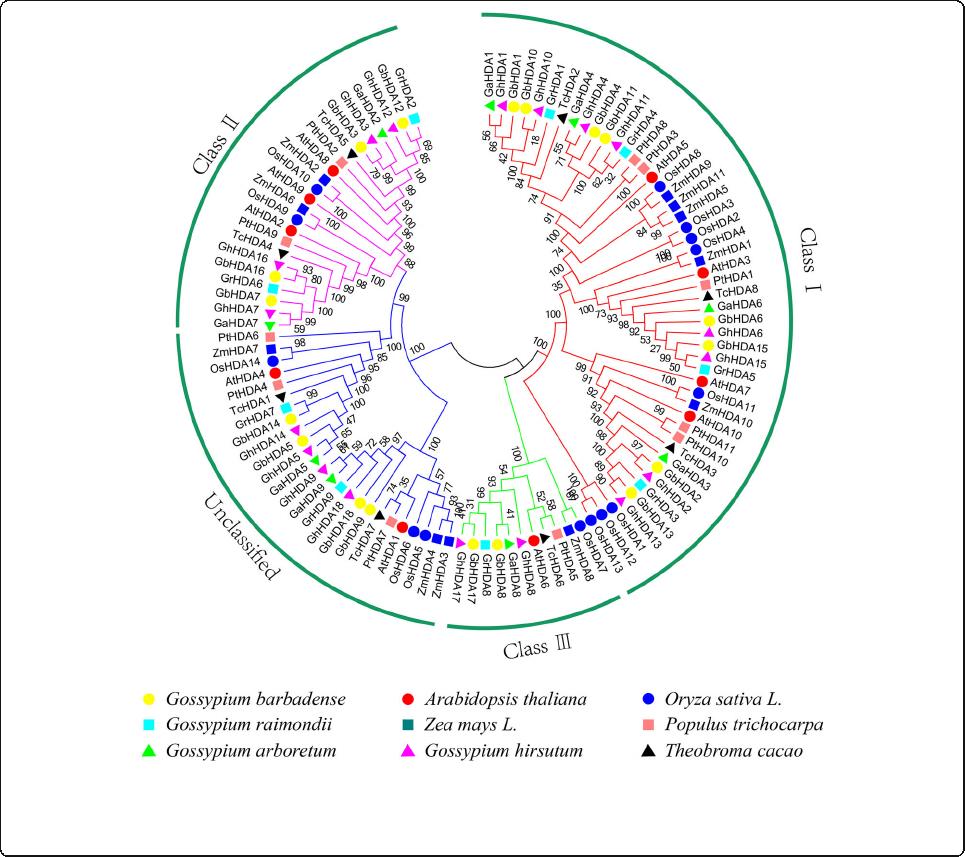
Phylogenetic analysis of the RPD3 gene family
A total of 108 identified RPD3 protein sequences from
G. raimondii (9), G. arboreum (9), G. hirsutum (18), G.
barbadense (18), A. thaliana (10), T. cacao (8), Oryza
sativa (14), Zea mays (11) and P. trichocarpa (11) were
employed to construct an unrooted phylogenetic tree
using the neighbor-joining method for investigating the
evolutionary relationships of RPD3 proteins. The RPD3
proteins were phylogenetically classified into 4 subfam-
ilies (Class I, Class II, Class III, and unclassified) accord-
ing to the formulated subfamilies in Arabidopsis [7]. The
Class I subgroup was the largest subfamily with 49
RPD3 genes, whereas the Class III subgroup has the few-
est members, only containing one gene in seven diploid
species genomes and two genes in two tetraploid cotton
genomes (Fig. 1). Among these four classes, each sub-
family contained RPD3 genes from all nine species, indi-
cating this gene family was relatively conserved in
different species during evolution.
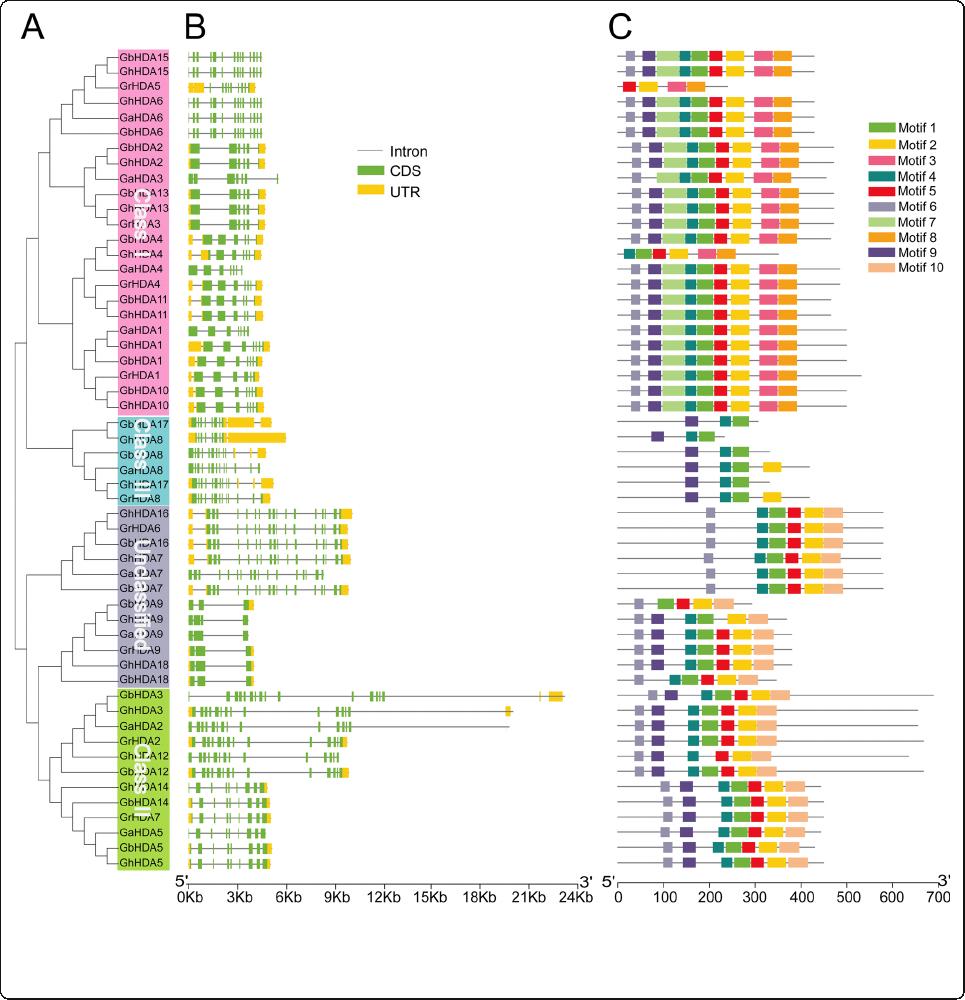
Exon-intron structure and conserved motif analysis
The domains of the RPD3 sequences in cotton were in-
vestigated and exhibited according to the results of the
SMART database using TBtools, revealing that all cotton
RPD3 genes contained a Hist_deacetyl domain (Add-
itional file 2: Table S2 and Additional file 3: Figure S1).
An unrooted phylogenetic tree with the predicted cotton
RPD3 genes was constructed (Fig. 2a), and then exon-
intron structure (Fig. 2b) and con served motifs (Fig. 2c)
were analyzed to better understand the similarity and
differences of cotton RPD3 members. The results
showed that the len gth of RPD3 cotton genes was rela-
tively conserved in Class I and Class III, but there were
twelve longer sequences in Class II and the unclassified
group. The RPD3 cotton genes included from 3 to 17
exons and most RPD3 genes (48/54) contained more
than five exons (Additional file 4: Table S3), which
might be associated with the diversification of their
functions. In terms of the distribution of motifs, most
RPD3 cotton genes belonging to the same subfamily
showed a similar motif composition, except in the un-
classified group (Fig. 2c). Most Class I subfamily mem-
bers contained 9 motifs, whereas GrHDA5 and GhHDA4
contained 4 and 6 motifs, respectively. Class III subfam-
ily genes had three or four motifs, and most Class II sub-
family members possessed 7 motifs, except for
GhHDA12 with 6 motifs. There were differences in the
exon-intron structure and motif arrangement among the
four categories, but they were highly conserved on the
same branches, indicati ng that the RPD3 members clas-
sified into the same branch might perform a relatively
conserved function in cotton growth and development.
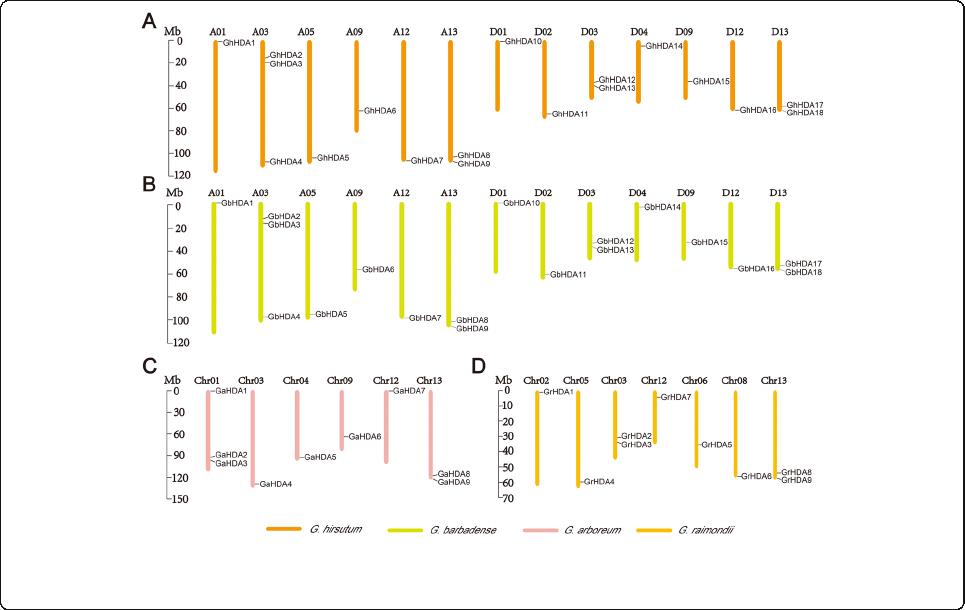
Chromosomal distribution, gene duplication and selection
pressure
The chromosomal distributions of GrRPD3 , GaRPD3,
GbRPD3, and GhRPD3 genes were visualized according
to the genomic position of 54 cotton RPD3 genes (Add-
itional file 5: Table S4 and Fig. 3). In G. hirsutum,18
GhRPD3 genes were unevenly mapped on 13
Table 1 Physicochemical parameters of 18 RPD3 genes in G. hirsutum
Name Gnen ID Protein Length Protein pI Protein MW (kD) Subcellular localization
GhHDA1 Ghir_A01G001410.1 499 4.9676 56.18 Nuclear
GhHDA2 Ghir_A03G007210.1 471 5.076 53.09 Nuclear
GhHDA3 Ghir_A03G008200.1 655 5.325 73.01 Cytoplasmic
GhHDA4 Ghir_A03G018610.1 351 4.4737 39.55 Cytoplasmic/Nuclear
GhHDA5 Ghir_A05G039610.1 449 6.9085 48.66 Mitochondrial/Chloroplast
GhHDA6 Ghir_A09G010210.1 429 4.8969 49.08 Cytoplasmic/Nuclear
GhHDA7 Ghir_A12G027820.1 574 6.3108 63.26 Cytoplasmic
GhHDA8 Ghir_A13G019980.1 232 6.5919 25.79 Plasma Membrane
GhHDA9 Ghir_A13G023460.1 368 5.3373 40.37 Cytoplasmic/Chloroplast
GhHDA10 Ghir_D01G001410.1 499 4.9676 56.26 Nuclear
GhHDA11 Ghir_D02G019970.1 465 5.1309 52.65 Nuclear
GhHDA12 Ghir_D03G010660.1 635 4.8889 71.02 Cytoplasmic
GhHDA13 Ghir_D03G011510.1 471 5.1489 53.06 Nuclear
GhHDA14 Ghir_D04G003510.1 443 6.9591 47.95 Chloroplast/Mitochondrial
GhHDA15 Ghir_D09G009940.1 429 4.8371 49.11 Cytoplasmic/Nuclear
GhHDA16 Ghir_D12G027930.1 579 6.1788 63.80 Cytoplasmic
GhHDA17 Ghir_D13G020760.1 331 8.6517 37.28 Plasma Membrane
GhHDA18 Ghir_D13G024090.1 380 5.648 41.63 Cytoplasmic
Zhang et al. BMC Genomics (2020) 21:643 Page 3 of 16
chromosomes. A03 contained the most GhRPD3
genes (3), whereas the other 12 chromoso mes only
contained one or two GhRPD3 genes (Fig. 3a). The
chrom osomal distribution of 18 GbRPD3 genes in G.
barbadense was similar to that of GhRPD3 gene s in
G. hirsutum (Fig. 3b). In G. arboreum,9GaRPD3
genes were unevenly located on 6 chromosomes.
Chr01 and Chr13 contained three and two GaRPD3
genes, respectively, and the other 4 chromosomes
contained only one GaRPD3 gene (F ig. 3c). In G. rai-
mondii, the chromosomal distribution of 9 GrRPD3
genes was highly consistent with the corresponding D
subgenome of G. hirsutum (Fig. 3d), sh owing con-
served numbers a nd chromosomal distribution of
RPD3 genes between diploid and tet raploid cotton
species. In addition, the lopsided chromosomal distri-
bution of the cotton RPD3 genes indicated that
genetic variation occurred during evolution. Notably,
most of the RPD3 genes were distributed on the o p-
posite ends of the chromosomes in f our cotton spe-
cies (Fig. 3).
In general, tandem and segmental duplication are
two of the main reasons for gene family generation
during evolution [21].Theanalysisofgeneduplica-
tion indicated that all RPD3 family members were
amplified o nly through segmental duplication (Add-
itional file 6: Table S5), suggesting that segmental du-
plication played a vital role in the evolution of the
RPD3 gen e family. The homologous gene pairs ob -
tained by collinearity analysis among RPD3 genes in
G. arboreum, G. raim ondii,andG. hirsutum were vi-
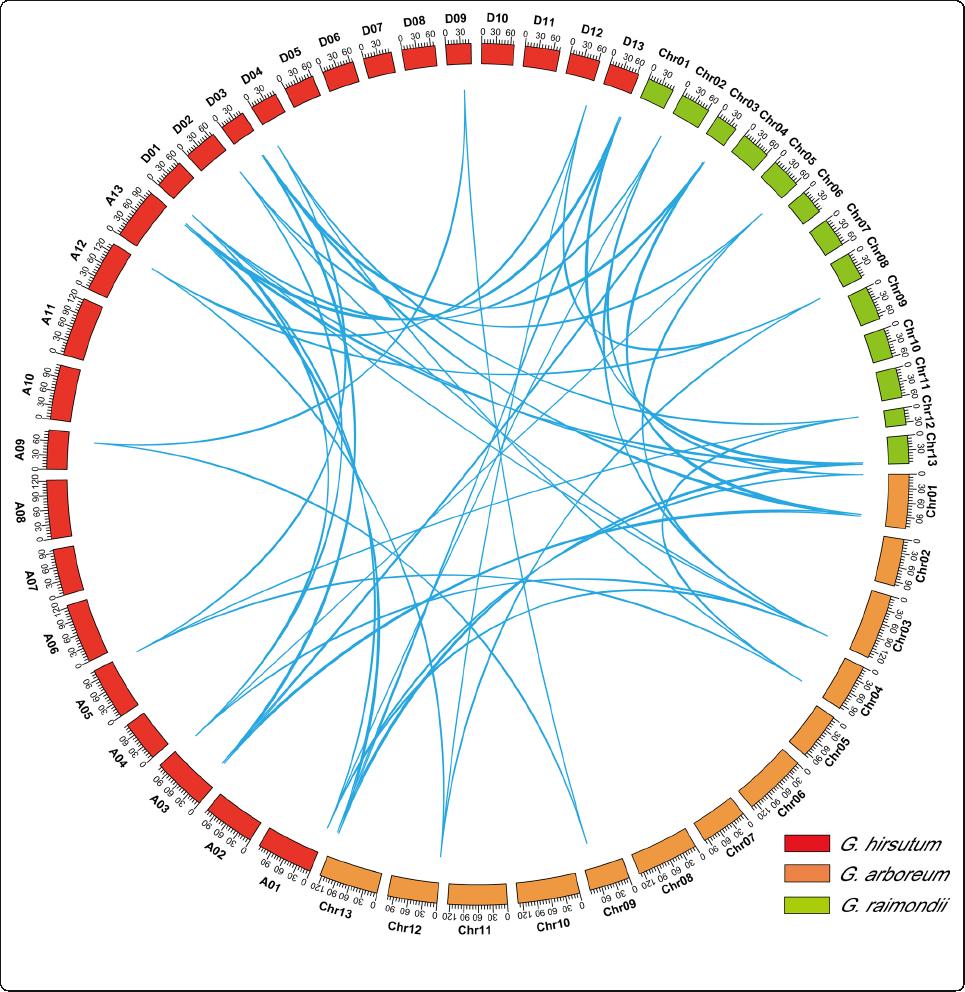
sualized using circular maps (Fig. 4). The Ka/Ks ratios
of most homologous gene pairs were lower than one,
indicating that purified selection was essential during
Fig. 1 Neighbor-joining phylogenetic tree of RPD3 gene family. The 108 predicted RPD3 proteins from G. hirsutum, G. arboreum, G. barbadense, G.
raimondii, A. thaliana, P. trichocarpa, T. cacao, Oryza sativa, and Zea mays were aligned using ClustalW, and the neighbor-joining (NJ) method was
used to construct this unrooted phylogenetic tree using MEGA 7.0 program with 1000 bootstrap repetitions. Four subfamilies are represented by
the different colored lines
Zhang et al. BMC Genomics (2020) 21:643 Page 4 of 16
the evolution of cotton RPD3 genes, whereas the Ka/Ks
ratios of two gene pairs (GhHDA2/GaHDA3 and
GhHDA6/GaHDA6)weremorethan1,suggestingthat
these two pairs might have experienced positive selection
pressure. The study also predicted the occurrence time of
segmentally duplicated RPD3 gene pairs by the formula
“t = Ks/2r” (r =2.6X10
− 9
)[22]. Except for the GhHDA6/
GaHDA6 gene pair, the other segmental duplication
events of three cotton species might have occurred 0.6 to
144.44 million years ago (MYA) with an average time of
18.39 million years ago (Additional file 6: Table S5).
Analysis of cis-elements in predicted promoter regions of
GhRPD3
To explore the possible regulatory functions of GhRPD3
genes under various envir onmental stresses and hor-
mone regulation pathways, the 2000-bp promoter re-
gions of 18 GhRPD3 genes were submitted to the
PlantCARE database for the identification of putative
stress-associated and plant hormone-related cis-
elements. A total of 9 kinds of elements related to plant
hormones, containing AuxRE-core (auxin), TGA-
element (auxin), P-box (gibberellin), TATC-box
Fig. 2 Phylogenetic relationships, exon-intron structure, and conversed motif analysis of cotton RPD3 genes. a A neighbor-joining phylogenetic
tree of 54 cotton RPD3 genes was generated using the MEGA7.0 program; (b) Exon-intron structure analysis of 54 cotton RPD3 genes. The UTRs,
exons, and introns are represented with yellow boxes, green boxes, and black lines, respectively; (c) The 10 conversed protein motifs of RPD3
genes are indicated by different colored boxes
Zhang et al. BMC Genomics (2020) 21:643 Page 5 of 16
(gibberellin), GARE- motif (gibberellin), CGTAC -motif
(MeJA), TGACG-motif (MeJA ), TCA-element (SA), and
ABRE (ABA), and 4 kinds of elements responding to
stresses, including TC-rich repeats (defense and stress
responsiveness), MBS (drought), WUN-motif (wound)
and LTR (cold stress), were predicted in the promoters
of GhRPD3 genes. As shown in Fig. 5, the promoters of
some GhRPD3 genes contained various hormone-
responsive and stress-responsive components, such as
GhHDA2 (2 MBS, 2 LTR, 2 TC-rich repeats, 1 GARE-
motif, 2 ABRE, 1 TGACG-motif) and GhHDA13 (1
MBS, 1 LTR, 1 TC-rich repeats, 1 AuxRE-core, 2 GARE-
motif, 1 TCA-element, 4 ABRE, 2 TGACG-motif).
Among the 18 GhRPD3 genes, there are large numbers
of light-responsive elements distributed in their pro-
moter regions (Additional file 7: Table S6). In addition,
MeJA-responsive and ABA-responsive elements are
more common than other hormone-related elements
(Additional file 8: Figure S2). These results revealed that
GhRPD3 genes might be involved in MeJA and ABA
hormone signaling pathways as well as response to en-
vironmental stresses.
Expression profiles of GhRPD3 genes in different tissues
and under different abiotic stresses
To understand the potential functions of GhPRD3 genes in
the growth and development of cotton, we studied their
expression in various cotton tissues, including the anther,
pistil, bract, sepal, petal, filament, torus, root, leaf, stem,
ovules, and fibers, using publicly available transcriptomic
data provided by Hu et al. [23]. Transcripts of all the
GhRPD3 genes were detected in at least three tissues with
fragments per kilobase million (FPKM) ≥ 1. Furthermore,
ten genes exhibited high expression levels in all selected tis-
sues (Additional file 9: Table S7). These results indicated
that GhRPD3 genes are widely expressed in both repro-
ductive organs and vegetative organs and thus might have
multiple biological functions. After log2-conversion of
FPKM values, the expression profiles of GhRPD3 genes in
differen t tissues are shown in Fig. 6a. Seven GhRPD3 genes
exhibited relatively high expression levels in at least eight
tissues (log2-transformed FPKM value≥2.6); in particular,
one pair of homologous genes (GhHDA1/GhHDA10)
showed a high expression level in all the tissues with a simi-
lar expression pattern. Nevertheless, three GhRPD3 genes
(GhHDA4, GhHDA14, GhHDA18) were expressed at rela-
tively low levels in at least eight tissues (log2-transformed
FPKM value< 1), of which GhHDA14 showed the lower
expression in all tissues except for the pistil. These
homologous gene pairs (GhHDA1/Gh HDA 10, GhHDA4/
GhHDA11, GhHDA2/GhHDA13, GhHDA6/GhHDA15,
GhHDA7/GhHDA16,andGhHDA9/GhHDA18)werelo-
cated on At and Dt subgenomes and exhibited similar ex-
pression patterns. For example, homologous gene pairs
Fig. 3 Chromosomal distribution of cotton RPD3 genes. a, b, c and d represent the chromosomal location of RPD3 genes from G. hirsutum (a), G.
barbadense (b), G. arboreum (c), and G. raimondii (d), respectively. The chromosome number is shown on the top of each chromosome. The scale
bars represent the length in mega bases (Mb)
Zhang et al. BMC Genomics (2020) 21:643 Page 6 of 16
(GhHDA4/GhHDA11 and GhHDA9/GhHDA18)showed
relative ly low expression in all twelve tissues. The
gene pair GhHDA2/GhHDA13 exhibited relatively
high expres sion in torus and ovule but relatively low
expression in petals (Fig. 6a).
Based on the analysis of cis-elements in promoter re-
gions and previous reports on RPD3 genes in other
plants, GhRPD3 gens might respond to abiotic stresses.
To test this hypothesis, we investigated the expression
characteristics of 18 GhRPD3 genes under cold, heat,
PEG, and salt treatments using available transcriptomic
data [23] (Fig. 6b). The expression of most GhRPD3
genes were induced by the four stresses to varying de-
grees. GhHDA1, GhHDA2, GhHDA6, GhHDA10,
GhHDA12, and GhHDA18 showed upregulated expres-
sion under four stress treatments. However, one gene
(GhHDA4) exhibited marked downregulation in the
presence of the four abiotic stresses. Some genes can re-
spond to one specific abiotic stress. For example, the ex-
pression of GhHDA13 and GhHDA16 was significantly
induced by PEG treatment. Four genes (GhHDA7,
GhHDA11, GhHDA5) showed upregulated expression
Fig. 4 RPD3 homologous gene pairs among G. arboreum, G. raimondii and G. hirsutum. Orange, blue and red represent chromosomes of G.
arboreum, G. raimondii and G. hirsutum, respectively
Zhang et al. BMC Genomics (2020) 21:643 Page 7 of 16
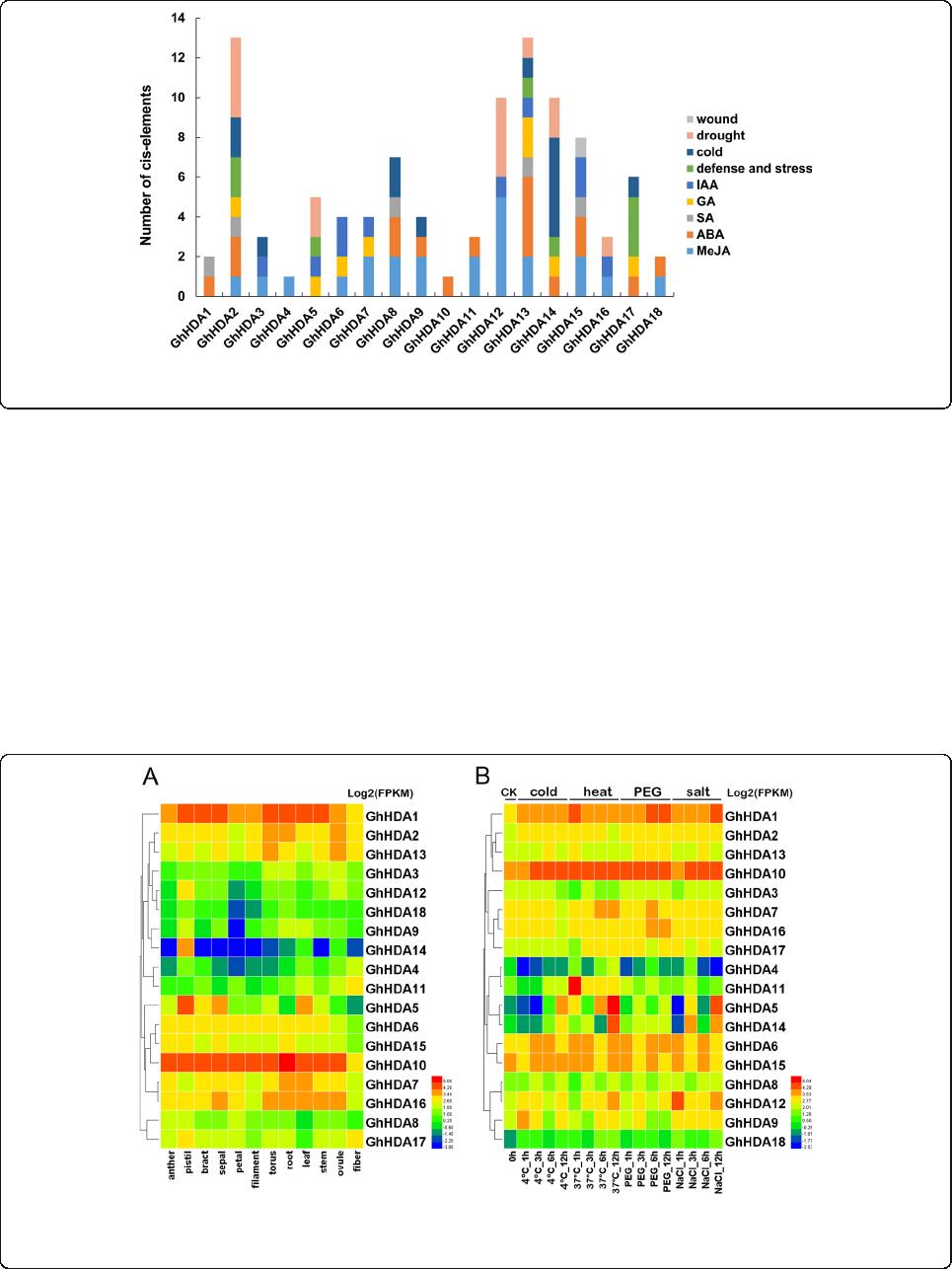
under heat treatment. The expression of GhHDA9 was
significantly upregulated under cold and salt treatments.
According to the results, we can conclude that GhRPD3
genes play an essential role in response to abiotic
stresses.
Characterization of GhRPD3 genes expression during
flower bud differentiation
To explore expression differences of GhRPD3 genes be-
tween early-maturity and late-maturity cottons during
flower bud differentiation, we selected nine genes show -
ing relatively high expression in floral organ tissues
(anther, pistil, bract, sepal, petal, filament and torus) for
qRT-PCR. The buds of an early-maturity variety
(CCRI50) and a late-maturity variety (GX11) from the
one-leaf to five-leaf stage were used for qRT-PCR (Fig. 7).
The results revealed that more than half of these genes
(5/9) possessed relatively higher expression in early-
maturity cotton compared with late-maturity cotton
during flower bud differentiation. GhHDA5 showed
marked differences at the two-leaf and three-leaf stages,
and these two stages were regarded as the important
period of flower bud differentiation. A homologous gene
pair (GhHDA6/GhHDA15) located on At and Dt
Fig. 5 Cis-elements of GhRPD3 genes in promoter regions. The numbers of different cis-elements are presented in the form of bar graphs, and
similar cis-elements are exhibited with the same colors
Fig. 6 Expression patterns of RPD3 genes in G. hirsutum. a and b represent the expression patterns of GhRPD3 genes in different tissues ( a) and
under four different abiotic stresses (b), respectively. Gene names are shown on the right. Scale bars on the right represent the log2-transformed
FPKM values of each gene
Zhang et al. BMC Genomics (2020) 21:643 Page 8 of 16
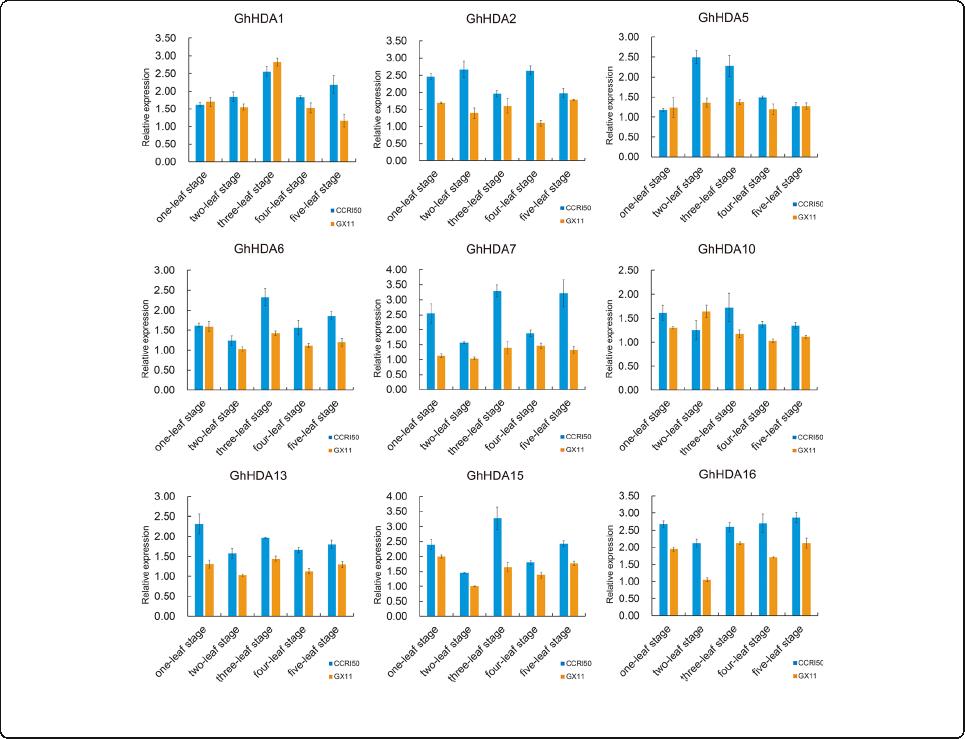
respectively, showed the same expression trend. Both of
them presented the highest expression at three-leaf stage
and then exhibited downregulated expression in next
two stages in CCRI50. In addition, all nine genes showed
relatively higher expression at the two-leaf or three-leaf
stage in CCRI50 compared with GX11. The results
showed that GhRPD3 genes are associated with the early
maturity of cotton.
Responses of GhRPD3 genes to MeJA and ABA treatment
MeJA and ABA play imp ortant roles in plant stress re-
sistance. To further explore the possible functions of
GhRPD3 genes, we selected the GhRPD3 genes
containing MeJA- and ABA-responsive elements in the
predicted promotors to analyze their expression charac-
teristics under MeJA and ABA treatment by qRT-PCR
(Figs. 8 and 9). Most GhRPD3 genes (8/13) were mark-
edly upregulated at 9 h after MeJA treatment. Three
genes (GhHDA7, GhHDA13,andGhHDA18)exhibited
significantly upregulated expression at three or more time
points, whereas four genes (GhHDA2, GhHDA8,
GhHDA9,andGhHDA11)showedmarkedtranscriptional
downregulation at least three time points after MeJA
treatment (Fig. 8). More than half of the GhRPD3 genes
(6/11) were significantly upregulated at 9 h after ABA
treatment. Three GhRPD3 genes (GhHDA14, GhHDA15,
and GhHDA18) showed relatively high expression at three
or more time points, whereas three GhRPD3 genes
(GhHDA10, GhHDA11,andGhHDA17)showedearly
downregulated and then upregulated expression patterns
under ABA treatment (Fig. 9). The results showed that the
exogenous application of MeJA and ABA significantly in-
duced the transcription of most GhRPD3 genes containing
MeJA-responsive and ABRE elements in their promoter
regions.
Discussion
Among the several histone modifications, histone acetyl-
ation plays an essential role in plant growth and devel-
opment [24]. Histone acetylation and deacetylation are
catalyzed by histone acetyltransferases (HATs) and his-
tone deacetylases (HDACs), respectively [20]. In plants,
HDACs are involved in a variety of biological processes
associated with plant growth and development [25].
Fig. 7 Expression levels of 9 GhRPD3 genes between CCRI50 and GX11. Blue and orange bar graphs indicate the expression of early-maturity
cotton (CCRI 50) and late-maturity cotton (GX11), respectively. The error bars show the standard deviation of three biological replicates
Zhang et al. BMC Genomics (2020) 21:643 Page 9 of 16
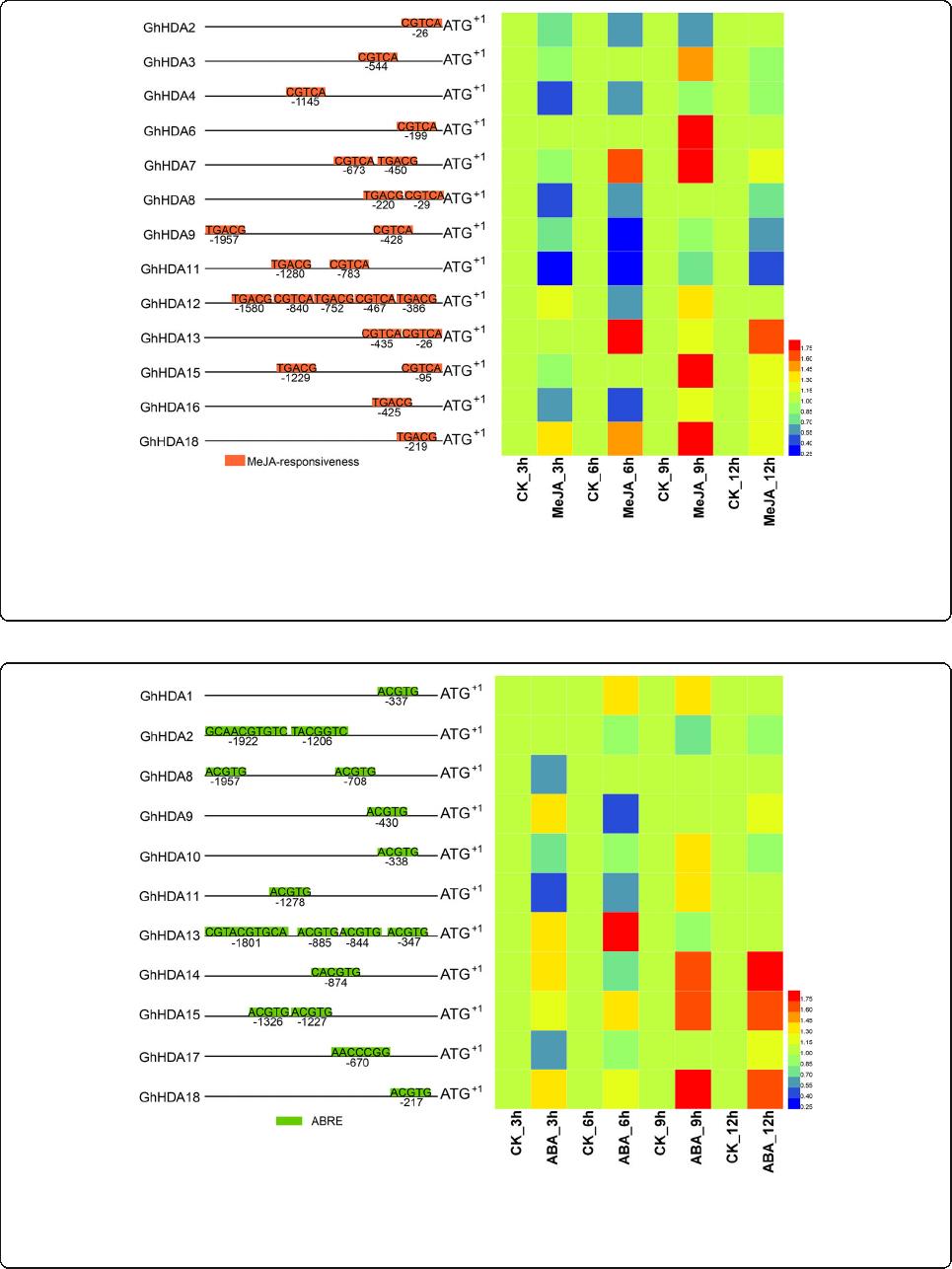
Fig. 8 Expression profiles of 13 GhRPD3 genes under MeJA treatment. Orange boxes represent the MeJA-responsive elements of 13 GhRPD3
genes in the promoter regions (left). The expression changes of 13 GhRPD3 genes under MeJA treatment are shown using a heatmap (right).
qRT-PCR was carried out with three technical and three biological replicates. Relative expression levels of each gene were calculated after
normalizing the expression level in CK (water) to 1.0
Fig. 9 Expression patterns of 11 GhRPD3 genes under ABA treatment. Green boxes represent the ABRE of 11 GhRPD3 genes in the promoter
regions (left). The expression changes of 11 GhRPD3 genes under ABA treatment are shown using a heatmap (right). qRT-PCR was conducted
with three technical and three biological replicates. Relative expression levels of each gene were calculated after normalizing the expression level
in CK (water) to 1.0
Zhang et al. BMC Genomics (2020) 21:643 Page 10 of 16
